
Not too long ago, I came across ranger; a package for the R project which can train and predict using random forests. In its source repository, issue #304 highlighted some wastefulness in training and prediction:
- Data is passed during training to a fast C++ library to construct the forest object (in C++).
- The object is then converted to an R object and passed to the user, and the C++ representation is destroyed (wasted).
- If the user needs predictions, they pass the R object back to the C++ library which reconstructs the C++ representation from the R object.
This “bad design” isn’t necessarily a problem. However, for algorithms like multiple imputation for missing data, the prediction step is used enough that it might be worth eliminating the waste.
With this in mind, and with with a goal of “better design”, I set about making literanger, which is now available on the official CRAN repository.
Not all features of the original ranger package are implemented in literanger, but it has performed 2x faster in multiple imputation tests, and it is now available to use in the popular mice R package, too.
Demonstration
For those who don’t know, random forests are one of the elder algorithms in machine learning, used to predict ’type’ (or class) given a variety of predictors.
A classic dataset used to demonstrate (and evaluate) machine learning algorithms is the MNIST data. This is a collection of 28x28 images of hand-written numbers from 0 to 9; e.g. here are three randomly selected images from the data set:
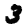 |
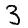 |
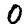 |
|---|
The whole data set of 60,000 images is available in .csv format here. Firstly, we load the data into R:
mnist_train_df <- read.csv('mnist_train.csv', header=FALSE)
mnist_test_df <- read.csv('mnist_test.csv', header=FALSE)
The data sets have 785 columns, with each row being one image. The first column is the correct number (label) for the image, the remaining columns are the intensity of each pixel. We can produce the images above via:
sample_image_A <- t(matrix(as.matrix(mnist_train_df[1234,-1]), nrow=28)) / 255
sample_image_B <- t(matrix(as.matrix(mnist_train_df[12345,-1]), nrow=28)) / 255
sample_image_C <- t(matrix(as.matrix(mnist_train_df[54321,-1]), nrow=28)) / 255
png::writePNG(1 - sample_image_A, 'mnist_sample_A.png')
png::writePNG(1 - sample_image_B, 'mnist_sample_B.png')
png::writePNG(1 - sample_image_C, 'mnist_sample_C.png')
To fit a random forest with 64 trees to the training data, the train
function is used, noting that the first column (the response) is called V1
and we want to classify each image as one of 10 possible types:
model_lr <- literanger::train(
data=mnist_train_df, response_name='V1', n_tree=64L, classification=TRUE
)
We can get predictions from the test data set via the predict generic:
predict_lr <- predict(model_lr, newdata=mnist_test_df)
Here are some examples of correct and incorrect predictions in the test dataset:
| MNIST |  |
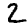 |
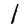 |
 |
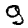 |
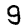 |
|---|---|---|---|---|---|---|
| Prediction | 7 | 2 | 1 | 4 | 8 | 8 |
The overall error rate was about 3.7%. Not great, really, but this is a highly naive approach to image classification! Advanced approaches like convolutional neural networks have performed a lot better.
Key features
The main features of literanger are:
- Training and prediction for classification and regression trees with a variety of splitting rules and customisations.
- Calculates the out-of-bag error during training.
- Faster training and prediction than ranger.
- Includes an additional prediction type used in multiple imputation via chained equations (see mice).
- Fast and compact serialization via cereal.
The speed difference can be demonstrated using the MNIST data and the microbenchmark package, e.g. training:
t_train <- microbenchmark::microbenchmark(
model_lr <- literanger::train(
data=mnist_train_df, response_name='V1',
n_tree=64L, classification=TRUE
),
model_rf <- ranger::ranger(
x=mnist_train_df[,-1], y=mnist_train_df[,1],
num.trees=64L, classification=TRUE
),
times=10L
)
# Unit: seconds
# min lq mean median uq max neval
# 8.163746 8.247235 8.441925 8.362481 8.710733 8.879759 10
# 8.887776 9.120844 9.388956 9.339751 9.594774 10.104999 10
And in prediction:
t_predict <- microbenchmark::microbenchmark(
predict_lr <- predict(model_lr, newdata=mnist_test_df),
predict_rf <- predict(model_rf, data=mnist_test_df),
times=10L
)
# Unit: milliseconds
# min lq mean median uq max neval
# 120.9096 122.1931 124.7303 125.4014 126.7077 128.3814 10
# 402.6253 410.2470 412.4108 413.2769 415.7943 416.5198 10
Serialization and deserialization of trained models is handled via
write_literanger and read_literanger respectively:
write_literanger(model_lr, 'foo.literanger')
model_lr_copy <- read_literanger('foo.literanger')
# use the trained model to predict again
predict(model_lr_copy, data=mnist_test_df)
Future plans
If there’s enough interest I would extend literanger to include the impurity measures and tree types that the original ranger package includes.
Acknowledgements
Obviously the package was derived from the original ranger package by Marvin H Wright:
Wright, M. N., & Ziegler, A. (2017a). ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. Journal of Statistical Software, 77, 1-17. doi:10.18637/jss.v077.i01.